Biochemically Active Metabolites of Gut Bacteria: Their Influence on Host Metabolism, Neurotransmission, and Immunity
| Received 17 Apr, 2025 |
Accepted 18 Jun, 2025 |
Published 19 Jun, 2025 |
The human gastrointestinal microbiota has emerged as a critical determinant of systemic health, functioning as a metabolically dynamic organ that produces a vast repertoire of bioactive compounds. This review presents a comprehensive synthesis of current insights into the role of gut microbiota-derived metabolites in regulating host metabolism, neurotransmission, and immune responses. Key microbial metabolites, such as Short-Chain Fatty Acids (SCFAs), secondary bile acids, Branched-Chain Amino Acids (BCAAs), tryptophan catabolites, and polyamines, modulate host physiology through diverse mechanisms including receptor-mediated signaling, epigenetic regulation, and neuroimmune interactions. The SCFAs influence metabolic homeostasis via G-protein-coupled receptors and histone deacetylase inhibition, while microbial transformation of bile acids affects lipid metabolism and endocrine signaling through FXR and TGR5 pathways. Additionally, gut microbes generate neuroactive molecules such as GABA, serotonin, and dopamine precursors that engage the gut-brain axis and impact cognitive and emotional states. Tryptophan metabolism along the kynurenine and indole pathways further connects microbial activity to neuroinflammation and immune tolerance via Aryl Hydrocarbon Receptor (AhR) and NMDA receptor modulation. Microbial-derived metabolites also shape mucosal and systemic immunity by promoting regulatory T-cell differentiation, modulating cytokine production, and activating pattern recognition receptors (PRRs) such as Toll-like and NOD-like receptors. Finally, parallels between gut-derived metabolic modulators and plant-based nanoparticle therapeutics highlight emerging translational avenues for microbial-mimetic interventions. By elucidating the biochemical networks underpinning host-microbe interactions, this review underscores the potential for microbiota-targeted strategies in the treatment of metabolic, neuropsychiatric, and inflammatory disorders.
| Copyright © 2025 Anih et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
The human gastrointestinal tract harbors a highly diverse microbial ecosystem composed of bacteria, archaea, viruses, and eukaryotic organisms collectively referred to as the gut microbiota. These microbes engage in symbiotic interactions with the host, influencing a wide range of physiological processes, including digestion, nutrient absorption, and immune development. Over the past decade, high-throughput sequencing and systems biology have provided compelling evidence that alterations in gut microbial composition and function are closely associated with diseases such as obesity, diabetes, and inflammatory bowel disease (IBD)1,2.
The functional impact of gut microbiota is largely mediated through its metabolic capacity to produce a wide spectrum of bioactive compounds. These metabolites, including Short-Chain Fatty Acids (SCFAs), bile acids, tryptophan derivatives, and neurotransmitter precursors, exert local and systemic effects on the host by modulating receptor signaling pathways, gene expression, and epigenetic modifications3.
The SCFAs, mainly acetate, propionate, and butyrate, are produced during the fermentation of dietary fibers by gut commensals and act through G-Protein-Coupled Receptors (GPCRs) such as GPR41 and GPR43 to regulate glucose and lipid metabolism. Beyond energy regulation, these SCFAs also influence inflammation and epithelial integrity, making them key players in host-microbiome communication4,5.
Additionally, microbial metabolites can transform host-derived bile acids into secondary bile acids, thereby influencing host lipid digestion and acting as signaling molecules through nuclear receptors like FXR and TGR55. Other metabolic byproducts, such as polyamines and Branched-Chain Amino Acids (BCAAs) contribute to mTOR pathway activation and energy metabolism, with implications in insulin sensitivity and metabolic health6.
This review synthesizes current knowledge on the molecular mechanisms by which gut microbiota-derived metabolites influence host metabolism, neurotransmission, and immunity. Understanding these biochemical pathways provides essential insight into the therapeutic modulation of the microbiome in chronic diseases.
GUT MICROBIAL METABOLITES AND HOST METABOLISM
Short-Chain Fatty Acids (SCFAs) and energy regulation: Short-chain fatty acids-primarily acetate, propionate, and butyrate-are the main products of anaerobic fermentation of dietary fibers by commensal gut bacteria such as Faecalibacterium prausnitzii, Roseburia spp., and Bacteroides spp. These metabolites play key roles in energy homeostasis by serving as substrates for ATP generation and by modulating metabolic signaling pathways in the liver, adipose tissue, and skeletal muscle7-9.
Mechanism of action: Short-Chain Fatty Acids (SCFAs)-mainly acetate, propionate, and butyrate-are produced through anaerobic fermentation of dietary fibers by gut commensals such as Faecalibacterium prausnitzii, Roseburia spp., and Bacteroides spp. These metabolites are central to energy regulation, serving both as substrates for host cellular metabolism and as signaling molecules7-9.
The SCFAs activate specific G-protein-coupled receptors, including GPR41 (FFAR3), GPR43 (FFAR2), and GPR109A, thereby modulating insulin sensitivity, lipid metabolism, and inflammatory responses. Additionally, butyrate inhibits Histone Deacetylases (HDACs), affecting gene transcription related to immune tolerance and epithelial maintenance7-9.
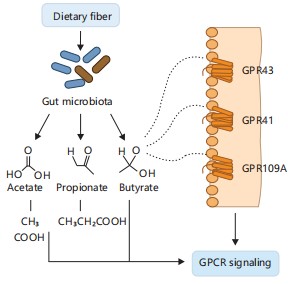
Figure 1 illustrates this mechanism by showing how gut microbiota ferment dietary fiber into SCFAs-acetate, propionate, and butyrate-which then activate G-protein-coupled receptors (GPR41, GPR43, GPR109A) on host cells7-9. This receptor activation modulates energy balance, immune signaling, and lipid metabolism. In addition, butyrate’s role as a Histone Deacetylase (HDAC) inhibitor is depicted, emphasizing its contribution to gene regulation and epithelial integrity.
|
Gut-derived SCFAs activate GPR41, GPR43, and GPR109A on host epithelial and immune cells, influencing metabolic homeostasis and inflammation. Butyrate also acts as an HDAC inhibitor, enhancing epithelial integrity and gene regulation7-9.
Acetate reaches peripheral tissues and contributes to cholesterol biosynthesis in the liver. Propionate supports hepatic gluconeogenesis, while butyrate is a key energy source for colonocytes and enhances mucosal barrier function7-9.
Bile acid biotransformation and lipid metabolism: Gut microbiota plays a pivotal role in modifying host-synthesized primary bile acids (cholic acid and chenodeoxycholic acid) into secondary bile acids (such as deoxycholic acid and lithocholic acid) through deconjugation and dehydroxylation reactions. These reactions are catalyzed by enzymes like bile salt hydrolases and 7α-dehydroxylases10.
These bile acid derivatives interact with host nuclear receptors such as farnesoid X receptor (FXR) and G-protein-coupled bile acid receptor 1 (TGR5), modulating lipid metabolism, glucose regulation, and inflammatory responses10.
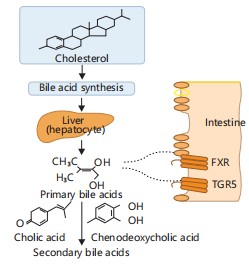
These metabolites interact with host nuclear receptors such as farnesoid X receptor (FXR) and G protein-coupled bile acid receptor 1 (TGR5), regulating glucose metabolism, lipid homeostasis, and inflammation. Figure 2 illustrates these processes, showing how microbial conversion of primary to secondary bile acids enables interaction with FXR and TGR5 receptors10. This signaling cascade integrates bile acid metabolism with host lipid regulation and immune modulation.
Gut microbiota converts primary bile acids to secondary bile acids, which activate host FXR and TGR5 receptors. This interaction regulates lipid digestion, bile acid homeostasis, and immune modulation10.
Impaired microbial bile acid metabolism has been linked to metabolic disorders, including Non-Alcoholic Fatty Liver Disease (NAFLD) and insulin resistance10.
Branched-Chain Amino Acids (BCAAs) and insulin resistance: Branched-Chain Amino Acids (BCAAs)-leucine, isoleucine, and valine-are essential amino acids whose circulating levels can be influenced by gut microbiota. Specific microbial populations can either produce or degrade BCAAs, thereby impacting host metabolic signaling11.
|
|
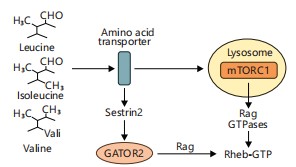
Elevated BCAA levels have been shown to activate the mTORC1 pathway, disrupting insulin signaling and contributing to insulin resistance, particularly in metabolic syndrome and type 2 diabetes11. Gut microbes such as Prevotella copri and Bacteroides vulgatus have been implicated in increasing systemic BCAA concentrations11. This pathway is visually summarized in Fig. 3, which depicts how leucine stimulates mTORC1 activation, linking microbial BCAA metabolism to host metabolic dysregulation. Reducing BCAA-producing microbes or increasing BCAA-catabolizing bacteria through dietary or probiotic interventions may thus offer therapeutic potential in managing insulin resistance and related metabolic disorders11.
Leucine activates mTORC1 via the Rag GTPase complex. Gut microbial activity that enhances BCAA availability contributes to metabolic dysregulation and insulin resistance11.
Modulating the gut microbial composition to favor BCAA catabolism may provide novel strategies to counteract metabolic diseases.
MICROBIOTA AND NEUROTRANSMISSION
Gut-brain axis and signaling: The gut-brain axis (GBA) is a bidirectional communication network linking the enteric and central nervous systems through neural, endocrine, immune, and metabolic pathways. Gut microbiota play a central role in modulating this axis, influencing brain development, behavior, and neuroinflammation12.
Microbial metabolites produced in the gut can impact the brain directly by crossing the blood-brain barrier or indirectly by activating the vagus nerve or immune mediators. Disruption of this microbial network, termed dysbiosis, has been associated with neurological conditions such as anxiety, depression, and neurodevelopmental disorders13,14.
|
In animal models, alteration of the gut microbiota has been shown to modify hippocampal neurotransmitter levels and behavioral responses, highlighting the influence of gut microbes on brain function15.
Production of neuroactive compounds: Gut microbes are capable of producing neuroactive molecules that modulate neurotransmission.
Gamma-Aminobutyric Acid (GABA) is synthesized by Lactobacillus and Bifidobacterium via glutamate decarboxylation. The GABA is the main inhibitory neurotransmitter in the central nervous system and modulates stress response and mood16.
Serotonin (5-HT) production is influenced by microbial regulation of enterochromaffin cell function and Tryptophan Hydroxylase 1 (TPH1), the rate-limiting enzyme for serotonin biosynthesis in the gut16.
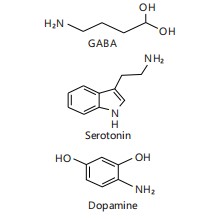
Figure 4 illustrates the chemical structures of GABA, serotonin, and dopamine, highlighting the role of gut microbes in generating these key neuroactive compounds16. This visual representation reinforces how microbial activity contributes to shaping the neurochemical environment of the host and supports gut-brain axis communication.
Gut microbiota modulates the synthesis of GABA, serotonin, and dopamine precursors. These molecules influence mood, cognition, and behavior by acting on the enteric nervous system or signaling to the CNS16.
Catecholamines such as dopamine and norepinephrine can be produced by bacteria, including Escherichia coli and Bacillus spp., impacting autonomic regulation and behavioral responses17.
These neuroactive compounds may signal locally via the enteric nervous system or systemically through neuroimmune pathways.
Tryptophan metabolism and kynurenine pathway: Tryptophan metabolism represents a critical link between the gut microbiota and host neurophysiology. The amino acid tryptophan can be metabolized via:
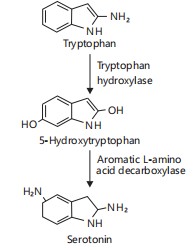
|
| • | Serotonin pathway, producing 5-HT | |
| • | Kynurenine pathway, resulting in neuroactive metabolites | |
| • | Indole pathway, catalyzed by gut bacteria, produces indole derivatives18 |
Gut microbes such as Clostridium sporogenes and Bacteroides spp. convert tryptophan into indole derivatives like indole-3-acetate and indole-3-aldehyde, which modulate epithelial barrier function and immune activity via the Aryl Hydrocarbon Receptor (AhR)18.
Meanwhile, host immune cells metabolize tryptophan into kynurenine through the enzyme indoleamine 2,3-dioxygenase (IDO), a pathway upregulated during inflammation. Kynurenine and its downstream products-quinolinic acid and kynurenic acid-can influence NMDA receptor activity, oxidative stress, and neuroinflammation19.
Figure 5 depicts the biosynthetic pathway of serotonin from tryptophan, involving enzymatic steps catalyzed by Tryptophan Hydroxylase (TPH1) and aromatic L-amino acid decarboxylase. This pathway illustrates how gut microbial modulation of host enzymes can influence serotonin availability and signaling19.
Tryptophan is processed by gut microbes into indole derivatives and by host cells via the kynurenine pathway. These metabolites affect neurotransmission and immune signaling through receptors such as AhR and NMDA19.
Altered balance between these metabolic routes has been associated with psychiatric disorders such as depression, schizophrenia, and cognitive decline19.
GUT MICROBIOTA AND IMMUNE MODULATION
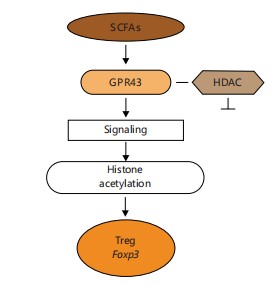
SCFAs and regulation of mucosal immunity: Short-Chain Fatty Acids (SCFAs), especially butyrate and propionate, are prominent immunomodulatory metabolites produced by gut microbes. These SCFAs influence immune homeostasis by promoting regulatory T cell (Treg) development, modulating cytokine profiles, and enhancing mucosal tolerance20.
Butyrate acts via two main mechanisms: Inhibition of Histone Deacetylases (HDACs) and activation of GPR43 on T cells and dendritic cells, leading to increased expression of Foxp3, a master regulator of Tregs21. This interaction is essential for the suppression of intestinal inflammation and the maintenance of epithelial integrity22.
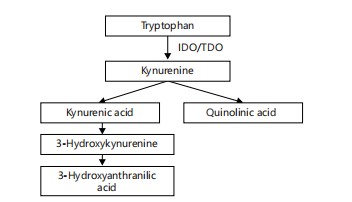
|
Figure 6 illustrates the kynurenine pathway of tryptophan metabolism, highlighting key intermediates such as kynurenic acid and quinolinic acid. These neuroactive and immunomodulatory metabolites are produced downstream of IDO activity and play distinct roles in modulating NMDA receptor activity, oxidative stress, and neuroinflammation22.
The SCFAs produced by gut bacteria stimulate regulatory T cell differentiation by activating GPR43 and inhibiting HDACs, leading to Foxp3 expression and immune tolerance in the gut22.
In addition to Tregs, SCFAs enhance the production of immunoglobulin A (IgA) by intestinal B cells and influence dendritic cell (DC) maturation toward a tolerogenic phenotype, reinforcing immune tolerance23.
Histamine and immune cell signaling: Certain microbes, such as Morganella morganii and Lactobacillus reuteri, generate histamine from histidine via histidine decarboxylase. This microbial histamine can signal through host histamine receptors H1R-H4R, differentially modulating inflammatory responses in the gut24.
Histamine can exert anti-inflammatory or pro-inflammatory effects depending on the receptor subtype it engages and the cellular context. For example, H2R activation on dendritic cells may suppress Th1 responses, while H1R engagement promotes allergic inflammation24.
Tryptophan-derived Indoles and the Aryl Hydrocarbon Receptor (AhR): Gut microbiota metabolizes dietary tryptophan into indole derivatives, such as indole-3-acetate and indole-3-aldehyde, which serve as ligands for the Aryl Hydrocarbon Receptor (AhR). The AhR is expressed in various immune cells and epithelial cells, where its activation regulates mucosal immunity and epithelial regeneration25.
The AhR signaling induces the production of Interleukin-22 (IL-22), a cytokine critical for epithelial barrier repair and antimicrobial peptide expression. It also supports the differentiation of intraepithelial lymphocytes and innate lymphoid cells that help maintain gut integrity25.
Figure 7 depicts how microbial metabolism of tryptophan through the kynurenine and indole pathways results in the production of neuroactive metabolites such as quinolinic acid and indole derivatives. These metabolites interact with NMDA receptors and the aryl hydrocarbon receptor (AhR), triggering microglial activation and neuroinflammatory responses. The figure highlights the dual role of gut-derived metabolites in both neurotransmission and neuroimmune regulation, offering insight into how microbial imbalance can contribute to psychiatric and neurodegenerative disorders25,26.
Microbial tryptophan metabolism generates indole derivatives that bind AhR. Activated AhR translocates to the nucleus, promoting IL-22 expression, enhancing epithelial barrier function, and antimicrobial defense25,26.
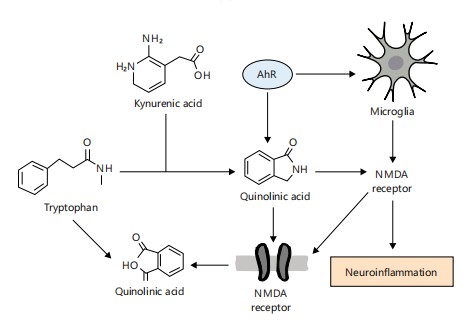
|
Defective AhR signaling has been linked to impaired barrier integrity and increased susceptibility to colitis and enteric infections26.
The host immune system senses microbes via pattern recognition receptors (PRRs) that recognize Microbial-Associated Molecular Patterns (MAMPs). These include:
Pattern recognition receptors (PRRs) and microbial sensing:
| • | TLRs: Toll-like receptors detect components such as lipopolysaccharides (TLR4), flagellin (TLR5), and unmethylated CpG DNA (TLR9)27 | |
| • | NLRs: NOD-like receptors, such as NOD2, detect intracellular bacterial peptidoglycans and activate immune responses through pathways like RIP2 and NF-κB27 |
Activation of PRRs triggers downstream signaling cascades that result in pro-inflammatory cytokine production, antimicrobial peptide release, and immune cell recruitment28.
As illustrated in Fig. 8, Short-Chain Fatty Acids (SCFAs) promote regulatory T cell (Treg) differentiation through a dual mechanism involving the activation of G-protein coupled receptor 43 (GPR43) and inhibition of Histone Deacetylases (HDACs). Activation of GPR43 by SCFAs initiates intracellular signaling that supports Treg expansion, while HDAC inhibition leads to enhanced expression of the transcription factor Foxp3, an essential marker of Treg identity and function. Together, these pathways reinforce mucosal immune tolerance and suppress inflammatory responses28,29.
The MAMPs like LPS, peptidoglycan, and CpG DNA are recognized by PRRs (e.g., TLRs, NOD2). These activate MyD88 or RIP2, leading to NF-κB-driven transcription of inflammatory and antimicrobial genes28,29.
Balanced PRR signaling supports immune homeostasis, while overactivation or defects in these pathways contribute to autoimmune disorders, chronic inflammation, and epithelial dysfunction29.
Microbial metabolite-induced biochemical shifts in host serum profiles: Gut microbial metabolites exert systemic effects on host physiology, including alterations in serum biochemical parameters such as glucose levels, urea, creatinine, liver enzymes, and electrolyte balance. These changes mirror findings from studies involving bioactive compounds derived from natural sources. For instance, administration of Azadirachta indica-derived silver nanoparticles in animal models led to significant modulation of serum transaminases, urea, and creatinine, suggesting possible overlap with microbial metabolite-mediated hepatic and renal responses.
|
Such biochemical shifts support the concept that gut-derived metabolites can interact with systemic organs beyond the intestine, modulating metabolic markers through receptor-mediated or epigenetic pathways. This has parallels with findings by Arowora et al.29, where distinct patterns in serum biochemical responses reflected the systemic absorption and activity of nanoparticle-bound phytochemicals.
Microbial metabolites such as butyrate and indoles are also known to influence electrolyte balance and blood pH regulation, an effect similarly observed in altered sodium and potassium profiles following nanoparticle administration. These shared mechanistic features underline the potential of both microbial metabolites and phyto-nanoconjugates as modulators of systemic homeostasis29.
Translational implications
From gut-derived metabolites to therapeutic nanoagents: The growing understanding of gut-derived biochemical modulators can be leveraged for developing novel therapeutic agents that mimic or enhance microbial signaling. Recent animal studies involving plant-based nanoparticles demonstrate comparable effects on immune and metabolic markers, indicating a promising translational pathway.
For example, gut microbial products such as SCFAs and polyphenolic metabolites influence hepatic enzyme activity and antioxidant responses-outcomes similarly mirrored in the serum biochemical landscape reported by Arowora et al.29. The observed reduction in ALT, AST, and bilirubin levels upon nanoparticle treatment reinforces the notion that biochemical modulators-whether microbial or synthetic-can induce hepatoprotective effects.
These mechanistic parallels are visually summarized in Fig. 9, which demonstrates how gut microbial metabolites and Azadirachta indica silver nanoparticles converge on similar physiological targets, particularly in liver and kidney function, as well as systemic immune modulation. The diagram underscores a shared ability to reduce key serum biomarkers like ALT, AST, urea, and creatinine, biomarkers that signal organ integrity and metabolic health.
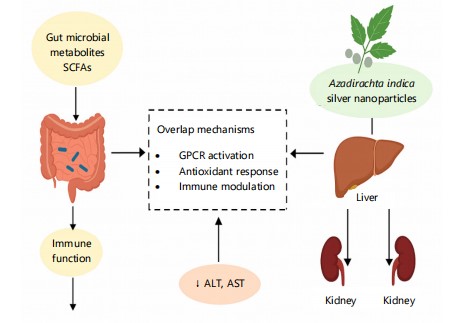
|
Figure 9 illustrates the comparative mechanisms by which gut microbial metabolites (e.g., SCFAs) and Azadirachta indica-derived silver nanoparticles exert systemic biochemical effects. Both pathways influence liver and kidney function as well as immune modulation through shared mechanisms such as GPCR activation, antioxidant response, and serum biochemical regulation (↓ALT, AST, urea, creatinine).
In summary, the study by Arowora et al.29 exemplifies how non-microbial agents can elicit host biochemical responses that resemble those of gut metabolites, providing a model system for further research into metabolite-mimicking therapeutics.
CONCLUSION
In conclusion, the gut microbiota functions as a metabolically active organ, orchestrating host physiological processes through its diverse array of bioactive metabolites. These microbial products influence key pathways in metabolism, neurotransmission, and immunity via complex biochemical and signaling mechanisms. The integration of microbial signals with host gene expression and epigenetic regulation underscores their systemic impact. Unraveling these interactions deepens our understanding of host-microbe symbiosis and its role in health and disease. This knowledge paves the way for microbiota-based diagnostics and therapeutic interventions. Continued research in this field holds immense promise for precision medicine and holistic disease management.
SIGNIFICANCE STATEMENT
Disruptions in gut microbial composition and function have been increasingly linked to metabolic, neurological, and immune-related disorders, yet the biochemical mechanisms underpinning these associations remain insufficiently understood. This study delineates how gut microbiota-derived metabolites, including short-chain fatty acids, bile acids, and tryptophan catabolites, modulate host metabolic regulation, neurotransmitter activity, and immune homeostasis through receptor signaling, epigenetic modification, and neuroimmune crosstalk. These findings offer critical insights into the molecular dialogues between the gut and host systems, advancing our understanding of the microbiota as a central regulator of systemic health. The implications are far-reaching, supporting the development of microbiome-targeted interventions, such as dietary modulation, probiotics, and metabolite-mimicking therapeutics, for personalized treatment of chronic metabolic, neuropsychiatric, and inflammatory diseases. This interdisciplinary synthesis opens new frontiers in microbiome science and translational medicine.
REFERENCES
- Nicholson, J.K., E. Holmes, J. Kinross, R. Burcelin, G. Gibson, W. Jia and S. Pettersson, 2012. Host-gut microbiota metabolic interactions. Science, 336: 1262-1267.
- Ridaura, V.K., J.J. Faith, F.E. Rey, J. Cheng and A.E. Duncan et al., 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science, 341.
- Velagapudi, V.R., R. Hezaveh, C.S. Reigstad, P. Gopalacharyulu and L. Yetukuri et al., 2010. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res., 51: 1101-1112.
- Liu, M., Y. Lu, G. Xue, L. Han and H. Jia et al., 2024. Role of short-chain fatty acids in host physiology. Anim. Models Exp. Med., 7: 641-652.
- Zhou, H., B. Yu, J. Sun, Z. Liu, H. Chen, L. Ge and D. Chen, 2021. Short-chain fatty acids can improve lipid and glucose metabolism independently of the pig gut microbiota. J. Anim. Sci. Biotechnol., 12.
- Wilkins, B.J. and M. Pack, 2013. Zebrafish models of human liver development and disease. Compr. Physiol., 3: 1213-1230.
- Cryan, J.F. and T.G. Dinan, 2012. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci., 13: 701-712.
- Carabotti, M., A. Scirocco, C. Severi and C. Severi, 2015. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol., 28: 203-209.
- Strandwitz, P., 2018. Neurotransmitter modulation by the gut microbiota. Brain Res., 1693: 128-133.
- Athnaiel, O., C. Ong and N.N. Knezevic, 2022. The role of kynurenine and its metabolites in comorbid chronic pain and depression. Metabolites, 12.
- Heijtz, R.D., S. Wang, F. Anuar, Y. Qian and B. Bjorkholm et al., 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A., 108: 3047-3052.
- Molska, M., K. Mruczyk, A. Cisek-Woźniak, W. Prokopowicz and P. Szydełko et al., 2024. The influence of intestinal microbiota on BDNF levels. Nutrients, 16.
- Turnbaugh, P.J., R.E. Ley, M.A. Mahowald, V. Magrini, E.R. Mardis and J.I. Gordon, 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444: 1027-1031.
- Wang, Y. and L.H. Kasper, 2014. The role of microbiome in central nervous system disorders. Brain Behav. Immun., 38: 1-12.
- Chatelier, E.L., T. Nielsen, J. Qin, E. Prifti and F. Hildebrand et al., 2013. Richness of human gut microbiome correlates with metabolic markers. Nature, 500: 541-546.
- Hoban, A.E., R.D. Moloney, A.V. Golubeva, K.A.M. Neufeld and O. O’Sullivan et al., 2016. Behavioural and neurochemical consequences of chronic gut microbiota depletion during adulthood in the rat. Neuroscience, 339: 463-477.
- Sampson, T.R. and S.K. Mazmanian, 2015. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe, 17: 565-576.
- Vyhlídalová, B., K. Krasulová, P. Pečinková, A. Marcalíková and R. Vrzal et al., 2020. Gut microbial catabolites of tryptophan are ligands and agonists of the aryl hydrocarbon receptor: A detailed characterization. Int. J. Mol. Sci., 21.
- Doña, I., N. Blanca-López, M.J. Torres, F. Gómez and J. Fernández et al., 2014. NSAID-induced urticaria/angioedema does not evolve into chronic urticaria: A 12-year follow-up study. Allergy, 69: 438-444.
- Arpaia, N., C. Campbell, X. Fan, S. Dikiy and J. van der Veeken et al., 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature, 504: 451-455.
- Furusawa, Y., Y. Obata, S. Fukuda, T.A. Endo and G. Nakato et al., 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature, 504: 446-450.
- Round, J.L. and S.K. Mazmanian, 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol., 9: 313-323.
- Belkaid, Y. and T.W. Hand, 2014. Role of the microbiota in immunity and inflammation. Cell, 157: 121-141.
- Kawai, T. and S. Akira, 2010. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol., 11: 373-384.
- Abreu, M.T., 2010. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol., 10: 131-144.
- Chen, G.Y. and G. Nuñez, 2010. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol., 10: 826-837.
- Spanogiannopoulos, P., E.N. Bess, R.N. Carmody and P.J. Turnbaugh, 2016. The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol., 14: 273-287.
- Tan, H.E., 2023. The microbiota-gut-brain axis in stress and depression. Front. Neurosci., 17.
- Arowora, K.A., K.C. Ugwuoke, M.A. Abah, A. Dauda and S.S. Ruyati, 2024. Evaluation of serum biochemical parameters in male Wistar rats administered with Azadirachta indica silver nanoparticles. Asian J. Sci. Technol. Eng. Art, 2: 462-475.
How to Cite this paper?
APA-7 Style
Anih,
D.C., Arowora,
A.K., Abah,
M.A., Ugwuoke,
K.C. (2025). Biochemically Active Metabolites of Gut Bacteria: Their Influence on Host Metabolism, Neurotransmission, and Immunity. Science International, 13(1), 46-57. https://doi.org/10.17311/sciintl.2025.46.57
ACS Style
Anih,
D.C.; Arowora,
A.K.; Abah,
M.A.; Ugwuoke,
K.C. Biochemically Active Metabolites of Gut Bacteria: Their Influence on Host Metabolism, Neurotransmission, and Immunity. Sci. Int 2025, 13, 46-57. https://doi.org/10.17311/sciintl.2025.46.57
AMA Style
Anih
DC, Arowora
AK, Abah
MA, Ugwuoke
KC. Biochemically Active Metabolites of Gut Bacteria: Their Influence on Host Metabolism, Neurotransmission, and Immunity. Science International. 2025; 13(1): 46-57. https://doi.org/10.17311/sciintl.2025.46.57
Chicago/Turabian Style
Anih, David, Chinonso, Adebisi Kayode Arowora, Moses Adondua Abah, and Kenneth Chinekwu Ugwuoke.
2025. "Biochemically Active Metabolites of Gut Bacteria: Their Influence on Host Metabolism, Neurotransmission, and Immunity" Science International 13, no. 1: 46-57. https://doi.org/10.17311/sciintl.2025.46.57

This work is licensed under a Creative Commons Attribution 4.0 International License.



