Detection and Phenotypic Characterization of Colistin-Resistant Bacteria in Water
| Received 18 Jan, 2023 |
Accepted 17 Apr, 2023 |
Published 03 Jul, 2023 |
Background and Objective: The emergence and dissemination of colistin-resistant Gram-negative bacteria strains are becoming a major global risk to human health, as colistin is one of the last-line antimicrobial agents against life-threatening infections. Thus, this study aimed at investigating the prevalence of colistin-resistant bacteria in various water sources. Materials and Methods: Samples were collected from taps connecting boreholes, stored rainwater, water distributors, or Ghana water company limited within Danfa, Otinibi and Cite-C, as well as from the Ayensu River. A total of 200 samples were collected from 5 different water sources, tap water (36 samples), borehole (96 samples), river (20 samples), rainwater (20 samples) and water distributors (28 samples). Results: A total of 200 water samples were collected from different sources with 46 aerobic Gram-negative bacteria isolated. Klebsiella species were the most isolated organism with Serratia marcescens being the least isolated. The 46% of the Gram-negative bacteria isolated were identified as resistant to colistin with minimum inhibitory concentration (MIC) results >2 µg mL‾1, while 54% were sensitive to colistin. Conclusion: Almost half of the organisms isolated during this study were resistant to colistin and a majority of these resistant organisms were obtained from borehole water samples. The prevalence of colistin-resistant bacteria was high in this study.
| Copyright © 2023 Osisiogu et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Gram-negative bacteria that are resistant to antibiotics are a serious worldwide health concern and are on the rise1. Chromosome-mediated routes of antibiotic resistance give rise to bacterial strains that are multi-drug resistant (MDR), extensively drug-resistant (EDR) and pan-drug resistant (PDR). Every year, these multidrug-resistant (MDR) bacteria cause illnesses that kill some 400,000 patients in Europe2,3.
Colistin is an antibiotic used to treat Gram-negative bacterial infections, of which most are Multi-Drug Resistant. The beta-lactamase enzyme action is a major factor discovered to be a cause of the rise of antibiotic resistance3.
Colistin is from the polymyxin E cationic polypeptide family, characterized by having a lipophilic fatty acyl side chain but its mechanism of killing is by targeting the cell membrane of the bacteria and by neutralizing the effects of endotoxins, these endotoxins of the Gram-negative bacteria (GNB) are found in the Lipid A site of the lipopolysaccharide (LPS) molecule but due to the high positive charge of colistin and its hydrophobic acyl chain, it interacts electrostatically with lipopolysaccharide (LPS) molecules and displaces its divalent cations, rupturing the cell membrane and increasing permeability and content leakage, ultimately causing cell death. Colistin was the best antibiotic in treating multi-drug resistant bacteria in 1959 until its effect led to nephrotoxicity and neurotoxicity in 1970 and was replaced with Aminoglycosides4. Colistin is now the only antibiotic available for treating infections caused by multidrug-resistant Gram-negative bacteria3 as drugs such as Carbapenems, Cephalosporins, quinolones, aminoglycosides and penicillin are failing. As of now, no new antibiotic has been discovered to replace colistin, therefore, it is important to keep monitoring MDR bacteria.
The spread of these MDR bacteria goes in a circular routine revolving around the essential habitats for their growth like our water bodies. Single contamination of any of these water sources by MDR organisms would lead to the widespread of such organisms among humans.
Water bodies are essential habitats for MDR organisms’ growth and a potential route for transmitting antibiotic-resistant organisms. Colistin resistance in bacteria has been proven recently to spread from cell to cell through plasmids in horizontal gene transfer known as plasmid-mediated colistin resistance mechanism coded as mobile colistin resistance genes (MCR). Findings have mcr-1 to mcr-10. Antibiotic resistance genes (ARGs), thus mcr2 discoveries have become rampant worldwide5. In 1947, a scientist from Japan, Koyama discovered Paenibacillus polymyxa (Colistinus) a sub-specie of a spore-forming soil bacterium belonging to the family of Polymyxin, which is why colistin is also known as Polymyxin E. The polymyxin family are polypeptide chains with hydrophobic fatty acid tails but are differentiated into five chemical groups, polymyxin A, B, C, D and E.
Antibiotic resistance became a major crisis in 1970, due to the increase in resistance of Gram-negative bacteria6. This has become a major crisis as Gram-negative bacterial infections are life-threatening and a common nosocomial infection as there is non-availability of effective alternative antibiotics and the emergence of multi-drug resistant (MDR) Gram-negative organisms, such as Escherichia coli, Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumonia worsens the situation.
Data gathering in Africa is limited on colistin resistance as the World Health Organization3 reports that antimicrobial resistance surveillance in Africa is hard as there are no viable medical data, unreliable laboratory capacity and scarce statistical information. Over the years, some African countries (Uganda, South Africa, Tunisia, Algeria, Nigeria, Rwanda and Egypt) have reported Colistin resistance from the community and hospital settings1. The spread of colistin-resistant bacteria is becoming more rampant globally and these resistant organisms are found in almost every aspect of the environment, such as in water sources, hospital settings, soil, poultry, etc. and are even termed “environmental contaminants”. Nosocomial infection of these organisms has become common and very difficult to treat, as the last resort antibiotic, colistin, is beginning to fail with resistance development towards it. Colistin traces are now being found in water, animal food production, farming soils and humans handling most animal activities.
The cautiousness of water treatment will increase as awareness of the incidence of colistin-resistant bacteria in communities grows. The investigations of colistin-resistant bacteria in a community will save both the patient and the laboratory time and energy on test investigation and save money as well for the patient.
This study is significant in that it would help reduce the rate at which colistin is administered wrongly, especially in communities that have demonstrated a heavy presence of colistin resistance traits and is a baseline for further studies into the molecular epidemiology of colistin-resistant genes in Accra and Ghana as a whole.
This study aimed at investigating the prevalence of colistin-resistant bacteria in various water sources such as rainwater, river, borehole water, stored tap water, direct tap water and water from water distributors in 4 communities.
MATERIALS AND METHODS
This survey was conducted from April to July, 2022.
Study design: The study is a cross-sectional study as it allows for the collection of data from a large pool of subjects and compares differences between groups.
Sampling technique: Samples were collected from rainwater, stored tap water, direct tap water, borehole, River Ayensu and from water distributors or Ghana water companies limited within Danfa, Otinibi and Cite-C, as well as from the Ayensu River. The samples were selected at random with informed consent obtained from the owners where necessary.
Study site: Samples were collected from the Danfa, Otinibi and Cite-C, communities and the Ayensu River. Ayensu River is located on the stretch from DM Farms through Ayensu River Estates to Danfa Royal Academy. The community is known as the new developing site with the majority of the indigenous population depending solely on Ghana water to meet their daily water needs whiles some also depend on the Ayensu River which is located at Danfa. The river is the main place for recreational activity for people in the surrounding community.
Study size and population: The sample size for the study was 200. Samples were collected from 5 different water sources, tap water (36 samples comprising both stored tap water and direct tap water), borehole (96 samples), river (20 samples), rainwater (20 samples) and water distributors (28 samples). These samples were sourced from four various communities, Danfa, Ayensu, Otinibi and Cite-C.
Laboratory process
Sample collection, handling and storage: Samples were collected into sterile 250 mL bottles from the different water sources used by the community inhabitants and immediately transported to the laboratory where the study was conducted upon sample arrival. The laboratory analysis was performed under strict aseptic conditions. Samples were preserved at -2∘C at least for 6 months after the initial study, for subsequent follow-ups and verification.
Water samples were randomly collected aseptically from five different sources, rainwater, stored and direct tap water, borehole, river and from water distributors in four distinct communities. The majority (52%) of the samples were collected in Danfa while sampling from Otinibi, Ayensu and Cite-C constituted 18, 16 and 14%, respectively.
Laboratory procedures: The syringe filter is a membrane filter that has a micropore size of 0.22 μm, a diameter of 13 mm and a hydrophilic and hydrophobic polytetrafluoroethylene (PTFE) to filter water sample Buxton et al.7. Using the syringe filtration method7, 50 mLs of water was filtered aseptically through 0.22 μm filter paper. The filter paper was then placed in Brain heart infusion (BHI) broth and incubated for 18-24 hrs. Growth from overnight incubation of BHI was sub-cultured onto both MacConkey agar to support most enteric bacterial growth and Salmonella Shigella agar for selective isolation of Salmonella and Shigella species and was incubated aerobically over a period of 18-24 hrs. The identity of each isolate was confirmed using a standard biochemical test after which they were examined for their susceptibility to colistin using both the Kirby Bauer Disk diffusion techniques and the broth microdilution technique8 for determining the minimum inhibition concentration (MIC).
Statistical analysis: The data were analyzed using SPSS (version 23.0) set at a 95% Confidence Interval and a p-value of <0.05 as statistically significant. The results were summarized by highlighting the percentage distribution in different categories and key associations were analyzed via the Chi-square test of association.
RESULTS AND DISCUSSION
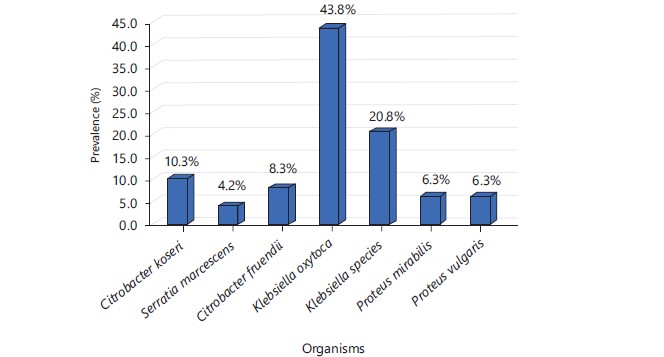
Results: During the study period, a total of 200 water samples collected from different sources (Fig. 1) were processed. A total of 48 aerobic Gram-negative bacteria were isolated from the water samples, with Klebsiella oxytoca and Klebsiella species accounting for 43.8 and 20.8%, respectively of the organisms isolated. The other organisms isolated include Citrobacter koseri (10.3%), Citrobacter fruendii (8.3%), Proteus vulgaris and Proteus mirabilis (6.3% each) and Serratia marcescens (4.2% each). The 46% of the isolates were phenotypically identified as colistin-resistant with MIC results >2 μg mL–1, while 54% were sensitive to colistin (Fig. 2). The high pattern of resistance observed in this study was similar to that which was observed in Nigeria where 17% of bacterial isolates from human and animals, including pigs, were resistant to colistin with resistant strains often associated with the mcr-1 gene9.
Demographics: Ayensu is a community located in the Greater Accra Region. It is surrounded by a water body, the Ayensu River. Indigens of the community are known for their farming activities. The major source of water in the community is the borehole water. As such, most houses in the area possess one of such amenities. Danfa on the other hand is a very large community comprising various ethnic groups and the indigenes are well known for their poultry and crop farming activities, as well as food distribution activities. The main sources of water the people depend on are tap water, borehole and water from water distributors. Cite-C is an estate-developed environment for most government officials and some titled military personnel. Over here, the main source of water is direct tap water as well as from water distributors. Otinibi is a community that is well known for its farming activities, the indigenes of this community are dependent on rainwater, borehole water and stored tap water as their main sources of water. The distribution of samples across the various communities and the frequency of sample types included in this study were described in Fig. 1.
Stored tap water for this study is water samples obtained from running taps but stored in household containers for future use by the indigenous.
The rise and spread of antibiotic-resistant bacteria is becoming a serious threat to public health globally. Several studies have recorded MDRs prevalence in water sources across the globe1, however, there is scarcity of data on colistin-resistance surveillance records in Ghana and most reports are on routine laboratory activities, not including colistin susceptibility and phenotypic testing. Colistin susceptibility testing has been carried out by the Clinical and Laboratory Standard Institute and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) since the establishment of a combined polymyxin breakpoints working group in March, 2016 that produced a standard operating code (BMD ISO 20776-1). As a result, the isolates from this study were confirmed using the Broth Micro Dilution Method. Kirby Bauer Technique on the other hand is a method that uses disc diffusion, however, it is not recommended for susceptibility testing against colistin10,11.

|

|
The Kirby Bauer Method of susceptibility testing is deemed inappropriate for determining colistin resistance due to the inability of colistin to properly diffuse through the agar media thus allowing bacteria to grow very close to the antibiotic disc and hence giving false resistance patterns. Although Kirby Bauer’s Method of susceptibility testing indicated 30% colistin resistance by the organisms isolated in the study, the MIC Method displayed a higher level of resistance (46%) to colistin (Fig. 2). Although the Kirby Bauer Method used in this study produced results that confirmed the report of Bardhan et al.12, it still goes to confirm the inability of colistin to diffuse properly through the agar media.
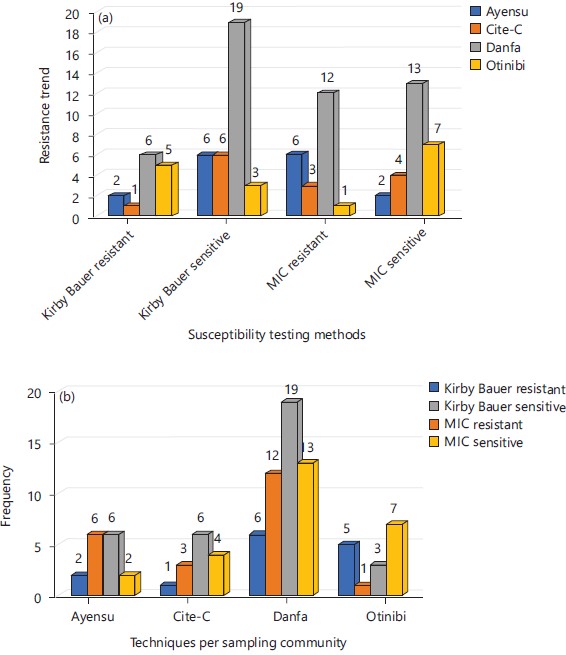
Comparing resistance trends across communities and between techniques: Figure 3(a-b) showed the resistance of isolates to colistin from the various communities. Samples from Danfa displayed the highest resistance to colistin. The microbroth dilution technique displayed a higher resistance profile compared to the Kirby Bauer disc diffusion techniques. This confirmed the report of van der Heijden et al.13 and Lo-Ten-Foe et al.14 that compared disc diffusion with E-test and broth microdilution for susceptibility testing.
|
Frequency of microbes isolated: The most prevalent organism isolated during this study is Klebsiella oxytoca (Fig. 4). Klebsiella species are generally known for their pathogenicity and virulence. Its presence in water is an indication of past contamination thus projecting long-term contamination of the water sources. This poses life-threatening harm to individuals who consume this water. The high prevalence of Klebsiella species in this study is contrary to the findings from South Ethopia15,16 that reported Escherichia coli as the most implicated in drinking water contamination.
Resistance distribution across communities using disk diffusion technique: Figure 5(a-b) depicted the resistance pattern across the various communities sampled using Kirby Bauer and micro broth dilution techniques, respectively.
In Ayensu, the resistant isolates out-numbered the sensitive isolates while in Danfa the resistant isolates almost equaled the sensitive isolates. The observation of a high prevalence of organisms in Danfa is a possible reflection of the large sampling carried out there and does not necessarily represent a high prevalence of microbes there compared to the other communities.
|

|
CONCLUSION
This finding is a report obtained from the phenotypic screening of isolates from various water sources used by communities for their daily activities against colistin sulfate antibiotics. In response to the influx of multi-drug resistant (MDR) Gram-negative bacteria, colistin has reemerged as a last-resort therapy for the treatment of life-threatening Gram-negative bacterial infections. However, this last line of protection against these deadly illnesses is seriously jeopardized by the emergence and rapid spread of colistin resistance. Almost half of the organisms isolated during this study were phenotypically resistant to colistin and a majority of these resistant organisms were obtained from borehole water samples. With this growing occurrence of resistance amongst Gram-negative organisms, Ghana must be precautious and well involved in advancing research relating to drug-resistant organisms. There is therefore the need for more research into colistin resistance and the launch of African-wide antibiotic stewardship.
RECOMMENDATIONS
In light of the above findings, we recommend that the molecular characterization of isolates involved in the spread of colistin resistance should be performed and documented. Also, routine laboratory phenotypic testing for colistin susceptibility should be established in hospital facilities in the country for surveillance. A campaign on colistin resistance should be endorsed by the government to educate the public and various communities on the rise and spread of colistin resistance by MDR Gram-negative bacteria and the consequences thereof.
SIGNIFICANCE STATEMENT
The general incidence of colistin resistance in human clinical isolates continues to be very low as a result of their relatively recent return to clinical procedures. However, due to the fast-rising usage of colistin in hospitals, which has enhanced selection pressure for resistance, there has been a striking rise in colistin-resistant bacteria in recent years. This warranted this study to investigate the presence of Colistin-resistant bacteria in water. The prevalence of phenotypically characterized colistin-resistant bacteria in water was found to be quite high. There is, therefore, the need for molecular characterization of colistin-resistant bacteria in water and a study to understand the mechanism of resistance.
ACKNOWLEDGEMENTS
The authors of this study are grateful to Mr. Akintade Akinola Ifeoluwa and Miss Njenka Tchanang Emmanuelle for their support during the laboratory phase of this study. We are also grateful to Mr. Kofi Mensah for his immense role in collecting samples from the Ayensu River.
REFERENCES
- Mousavi, S.M., S. Babakhani, L. Moradi, S. Karami and M. Shahbandeh et al., 2021. Bacteriophage as a novel therapeutic weapon for killing colistin-resistant multi-drug-resistant and extensively drug-resistant gram-negative bacteria. Curr. Microbiol., 78: 4023-4036.
- Nation, R.L. and J. Li, 2009. Colistin in the 21st century. Curr. Opin. Infect. Dis., 22: 535-543.
- Aruhomukama, D., I. Sserwadda and G. Mboowa, 2019. Investigating colistin drug resistance: The role of high-throughput sequencing and bioinformatics. F1000Research, 8.
- Bialvaei, A.Z. and H.S. Kafil, 2015. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin., 31: 707-721.
- Luo, Q., Y. Wang and Y. Xiao, 2020. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosaf. Health, 2: 71-78.
- Ahmed, M.A.E.G.E.S., L.L. Zhong, C. Shen, Y. Yang, Y. Doi and G.B. Tian, 2020. Colistin and its role in the era of antibiotic resistance: An extended review (2000-2019). Emerging Microbes Infect., 9: 868-885.
- Buxton, A., J. Groombridge and R. Griffiths, 2018. Comparison of two citizen scientist methods for collecting pond water samples for environmental DNA studies. Citiz. Sci.: Theory and Pract., 3: 2.
- Simar, S., D. Sibley, D. Ashcraft and G. Pankey, 2017. Colistin and polymyxin B minimal inhibitory concentrations determined by etest found unreliable for gram-negative bacilli. Ochsner J., 17: 239-242.
- Ngbede, E.O., A. Poudel, A. Kalalah, Y. Yang and F. Adekanmbi et al., 2020. Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int. J. Antimicrob. Agents, 56: 106108.
- Halaby, T., N. Al Naiemi, J. Kluytmans, J. van der Palen and C.M.J.E. Vandenbroucke-Grauls, 2013. Emergence of colistin resistance in Enterobacteriaceae after the introduction of selective digestive tract decontamination in an intensive care unit. Antimicrob. Agents Chemother., 57: 3224-3229.
- Tan, T.Y. and L.S.Y. Ng, 2006. Comparison of three standardized disc susceptibility testing methods for colistin. J. Antimicrob. Chemother., 58: 864-867.
- Bardhan, T., M. Chakraborty and B. Bhattacharjee, 2020. Prevalence of colistin-resistant, carbapenem-hydrolyzing proteobacteria in hospital water bodies and out-falls of West Bengal, India. Int. J. Environ. Res. Public Health, 17: 1007.
- van der Heijden, I.M., A.S. Levin, E.H. de Pedri, L. Fung and F. Rossi et al., 2007. Comparison of disc diffusion, Etest and broth microdilution for testing susceptibility of carbapenem-resistant P. aeruginosa to polymyxins. Ann. Clin. Microbiol. Antimicrob., 6: 8.
- Lo-Ten-Foe, J.R., A.M.G.A. de Smet, B.M.W. Diederen, J.A.J.W. Kluytmans and P.H.J. van Keulen, 2007. Comparative evaluation of the VITEK2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob. Agents Chemother., 51: 3726-3730.
- Dhengesu, D., H. Lemma, L. Asefa and D. Tilahun, 2022. Antimicrobial resistance profile of Enterobacteriaceae and drinking water quality among households in Bule Hora Town, South Ethiopia. Risk Manage. Healthcare Policy, 15: 1569-1580.
- Yenew, C., M. Kebede and M. Mulate, 2022. Drinking water antimicrobial resistance enteric bacterial load and public health risk in Northwest, Ethiopia: A laboratory-based cross-sectional study. Ethiop. Med. J., 60.
How to Cite this paper?
APA-7 Style
Osisiogu,
E.U., Appiah,
C.A., Mahmoud,
F.C., Bawa,
F.K., Nattah,
E.M. (2023). Detection and Phenotypic Characterization of Colistin-Resistant Bacteria in Water. Science International, 11(1), 9-17. https://doi.org/10.17311/sciintl.2023.09.17
ACS Style
Osisiogu,
E.U.; Appiah,
C.A.; Mahmoud,
F.C.; Bawa,
F.K.; Nattah,
E.M. Detection and Phenotypic Characterization of Colistin-Resistant Bacteria in Water. Sci. Int 2023, 11, 9-17. https://doi.org/10.17311/sciintl.2023.09.17
AMA Style
Osisiogu
EU, Appiah
CA, Mahmoud
FC, Bawa
FK, Nattah
EM. Detection and Phenotypic Characterization of Colistin-Resistant Bacteria in Water. Science International. 2023; 11(1): 9-17. https://doi.org/10.17311/sciintl.2023.09.17
Chicago/Turabian Style
Osisiogu, Emmanuel, Udochukwu, Calebina Ayeyi Appiah, Fareeda Ceeta Mahmoud, Flavia Kaduni Bawa, and Emmanuel Mawuli Nattah.
2023. "Detection and Phenotypic Characterization of Colistin-Resistant Bacteria in Water" Science International 11, no. 1: 9-17. https://doi.org/10.17311/sciintl.2023.09.17

This work is licensed under a Creative Commons Attribution 4.0 International License.



